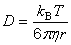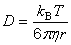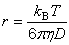|
| Constructors | public ImmunoChemistry() | |
| Diffusion coefficient | public static double diffusionCoefficient(double effectiveRadius, double viscosity, double temperature) | |
| public static double diffusionCoefficient(double molWeight, double specVol, double viscosity, double temperature) | ||
| Diffusion controlled reaction rate | public static double diffusionControlledRate(double diffCoeffA, double diffCoeffB, double radiusA, double radiusB, double fractA, double fractB) | |
| Planar Diffusion | public static double oneDimensionalDiffusion(double diffCoeff, double zeroDistConcn, double distance, double time) | |
| Molecular radius | public static double molecularRadius(double molWeight, double specVol) | |
| public static double molecularRadius(double molWeight) | ||
| public static double effectiveRadius(double diffusionCoefficient, double viscosity, double temperature) | ||
| Surface concentrations | Surface molar concentration | public static double surfaceMolarConcn(double effectiveRadius) |
| public static double surfaceMolarConcn(double molWeight, double specVolume) | ||
| Surface number concentration | public static double surfaceNumberConcn(effectiveRadius) | |
| public static double surfaceNumberConcn(double molWeight, double specVolume) | ||
| Equivalent molar volume concentration | public static double equivalentVolumeConcn(double effectiveRadius, double area, double volume) | |
| public static double equivalentVolumeConcn(double molWeight, double area, double volume, double specVolume) | ||
| public static double convertSurfaceToVolumeConcn(double surfaceConcn, double area, double volume) | ||
| Molecular weights | IgG1 | public static double getMolWeightIgG1() |
| IgG2 | public static double getMolWeightIgG2() | |
| IgG3 | public static double getMolWeightIgG3() | |
| IgG4 | public static double getMolWeightIgG4() | |
| IgM | public static double getMolWeightIgM() | |
| IgA1 | public static double getMolWeightIgA1() | |
| IgA2 | public static double getMolWeightIgA2() | |
| IgD | public static double getMolWeightIgD() | |
| IgE | public static double getMolWeightIgE() | |